VII. |
Human Genomic Melting Map being Included in the Ensembl Genome Browser as a DAS Source |
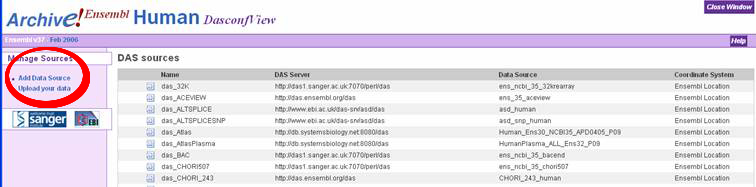
How to add the melting map DAS server
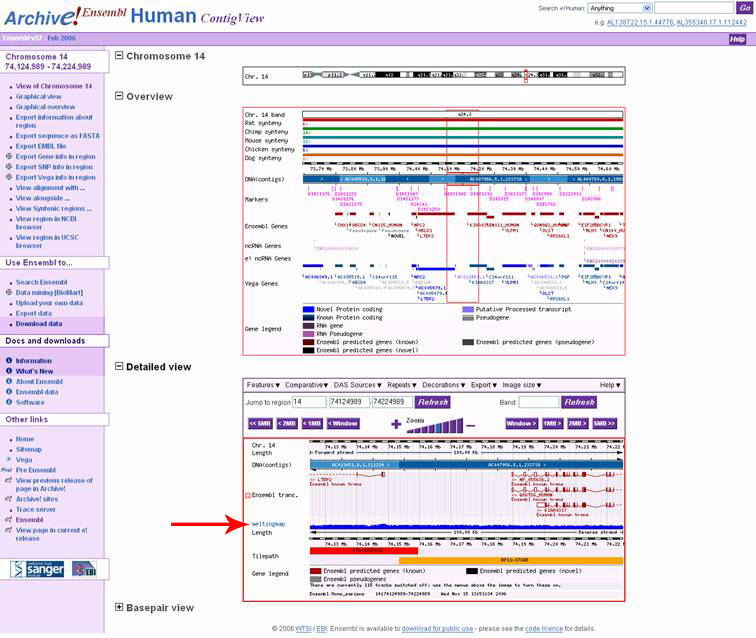
The melting map is available for browsing using an earlier version of Ensembl
containing the hg17 genome with annotations, as this is the genome release
for which we have reported calculations. The present version will become available
later. The process of adding the server to Ensembl is described step-by-step
below.
The February 2006 Version 37 of Ensembl is the one needed for browsing the
melting map, and the link for this can be found in the Ensembl Archives, or
alternatively by clicking this link:
http://feb2006.archive.ensembl.org/Homo_sapiens/index.html

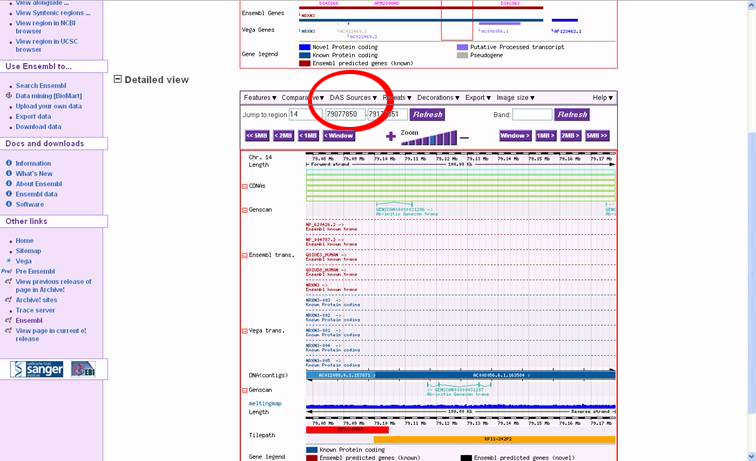
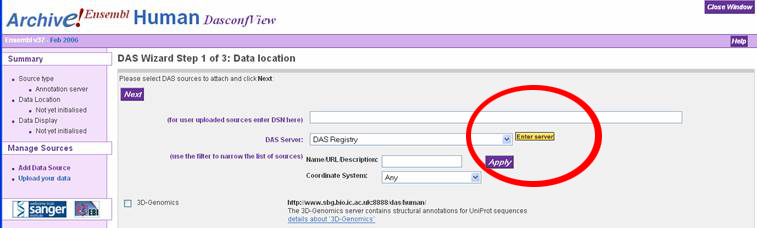
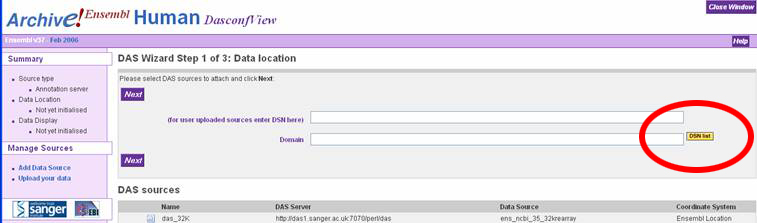
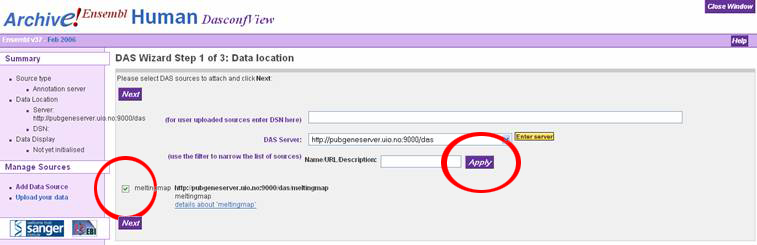
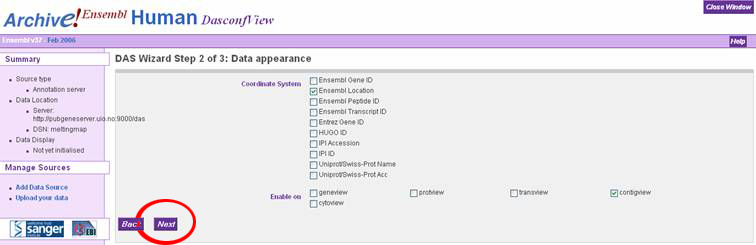
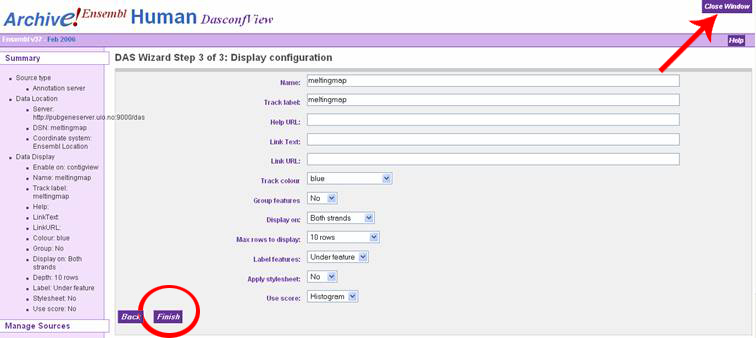
In the "DAS Wizard Step 3 of 3: Display configuration" window, the user can
specify a name for the DAS source, or just accept the defaults. Click the dropdown
box called "Use score" and select "Histogram" to display the melting map properly.
Click 'Finish', then the it is very important to click the 'Close Window' 'button
in the upper right corner of the window. This will result in an automatic
refresh of the ContigView page with the melting map displayed in the bottom
of the 'Detail View' part of the page. Unfortunately, there is no way to improve
the graphics further with Ensembls graphics handling ability with this version
of Ensembl. The upcoming calculation of hg18 will remedy this to some extent.